There is an increasing demand for biodiversity data, information and knowledge by the public and stakeholders. For this information to be widely available the existing technical barriers for interaction of e-infrastructures have to be opened. The pro-iBiosphere project has been launched for a period of two years (September 1st, 2012 to August 31st, 2014), with the goal of addressing technical and semantic interoperability challenges and preparing the ground for the creation of a system for intelligent management of biodiversity knowledge which will improve the present system of taxonomic literature.
One of the objectives of pro-iBiosphere is to promote and increase cooperation between the major biodiversity projects, initiatives and platforms at EU and global level (for additional objectives please seehttp://wiki.pro-ibiosphere.eu/wiki/Explaining_pro-iBiosphere).The persisting traditional workflow for producing taxonomic information, such as Floras and Faunas, is a very time-consuming process for most institutes (i.e. Natural History Museums, Herbaria, Botanic Gardens, etc.). These institutions have the responsibility to make sure that data are generated, curated and disseminated, and that technical operations are working adequately. An Open Biodiversity Knowledge Management System would not only facilitate the open access of taxonomic data, but it would create synergies with other initiatives / projects and through this allow to link taxonomic data in a wider context. These linkages will also offer many means of enhancement for the authors of this taxonomic information.
Examples of European e-infrastructure initiatives include EDIT, BHL-E, BioCASE, ViBRANT, PLAZI, biowikifarm, Pensoft, diversity workbench, BioVeL, and PESI. Over the last years a great number of software platforms have been developed with different functional scopes, content scopes, strengths and weaknesses. Biodiversity informatics infrastructures were often developed as database-driven web-applications which were later equipped with services for machine to machine communication. True interoperability is still hindered by a lack of data standards, standard protocols and solid service implementations.
Pro-iBiosphere will conduct four pilot studies (http://wiki.pro-ibiosphere.eu/wiki/Pilots) and organize six meetings with stakeholders (http://wiki.pro-ibiosphere.eu/wiki/Meetings). ; The FUB-BGBM and Plazi will be responsible for the pilot on "Interoperability model between PLAZI and the EDIT Platform for Cybertaxonomy based on transformations between XML-repositories and CDM-stores". Main focus will be optimized information- and workflows associated to mobilization, storage and publication of data from biodiversity literature. This includes the review of the existing system, working interfaces, successfully shared standards. The pilot implementation will be used as a proof of concept and establish a transformation pipeline between data from PLAZI (XML-based repositories) and the CDM-stores of the EDIT Platform for Cybertaxonomy (highly granular object-oriented data stores). The pilot will provide both human readable and web-service access to the Plazi data.
With the aim of identifying issues hindering true interoperability (e.g. lack of standards and different conceptual models etc.), a workshops on "How to improve technical cooperation and interoperability at the e-infrastructure level" will take place on October 8-11, 2013, in Berlin (http://wiki.pro-ibiosphere.eu/wiki/Pro-iBiosphere_stakeholders_meetings_and_workshops). The workshop will bring together IT- experts and representatives of e-infrastructures, data providers, and scientific users to identify the technical constraints and problems in the interoperability between platforms. Main outputs of the workshop are: (i) to specify workflows for importing mark-up documents into e-platforms, and (ii) a proposal of optimal solutions that will account the "different start" approach implied by the various stakeholder groups.
Eckert, S., Kelbert, P., Güntsch, A., Berendsohn, W. G. Botanischer Garten und Botanisches Museum Berlin-Dahlem Freie Universität Berlin (FUB-BGBM) & Sierra, S. Naturalis Biodiversity Center Leiden
In the field of biodiversity, the traditional workflow for producing core taxonomic information, such as Floras and Faunas, has not changed much over the years. The process is time-consuming process usually performed by individual specialists. Legacy (i.e. existing) literature is still the foundation and starting point for taxonomic studies. This literature contains all relevant data for a certain taxon, such as morphological characters, geographic distribution, taxonomic status, but also information that is needed to locate the physical specimens that were used to describe the taxon in the past. However, accessibility to legacy literature is uneven and a major drag on the pace of biodiversity research.
Since 2007, ten major biodiversity libraries have collaborated in digitising a large body of biodiversity literature (with a focus on English language literature) in an open access milieu via the Biodiversity Heritage Library (BHL) project. Since 2009, the eContentplus project Biodiversity Heritage Library for Europe (BHL-Europe) has achieved substantial progress in coordinating the digitisation of biodiversity literature in the EU. In the recent years, BHL has become a real global initiative: The Global Biodiversity Heritage Library (gBHL) is now a cooperative network of autonomous decentralised members operating programs and projects to make biodiversity literature more widely available. The current gBHL partner projects are: BHL, BHL-Europe, BHL-China, BHL-Australia, the BHL-SciELO Network (Brazil) and the BHL Arabic node organised by the Bibliotheca Alexandrina (Egypt). At the time of this writing (December 2012), more than 39 million pages from more than 109,000 books and journals are accessible online in an open access Creative Commons framework to a wide spectrum of end-users. This significantly facilitates the process of discovering and accessing literature that is relevant for taxonomic studies.
Currently, BHL content is available through various online portals (e.g. www.biodiversitylibrary.org, http://citebank.org/, www.bhl-europe.eu). Currently, most literature relevant to taxonomic studies are still manually extracted from this corpus of digitised literature. Semantic enhancements of the digitized literature could make the information in the literature even more accessible to researchers as well as amenable to searches and sophisticated queries. Taxonomic literature is ideal for (semi-)automatic enhancements as the description of the world’s biodiversity is a highly standardized process with a distinct language and format. Taxonomic treatments, for example, are a key structural element in taxonomic publications, where the various taxa (families, genera, species, etc.) are described.
Binomial species names are another key element that is highly standardized in taxonomic literature. Improving the power of users to search electronically for species names that appear in the literature has been a major focus for data enhancement in the various gBHL projects in recent years. The application of a name finding algorithm based on the OCRed (OCR = Optical Character Recognition) page images of the digitised literature facilitate the search for binomial species names. A search term expansion for common names and synonyms of scientific names further facilitates the search for animals and plants described in the literature. However, these are just first steps. Large-scale data mining of taxonomic literature is still very difficult, but further improvements in the structure of digitised taxonomic literature to facilitate increasingly sophisticated searches and queries are on the horizon. The pro-iBiosphere project will help identify current gaps in the process and recommend priorities for further development of tools and services to optimize the semantic mark-up of taxonomic literature. The project also helps to identify the necessary steps to improve the integration of biodiversity literature into a biodiversity knowledge management system (i-Biosphere). Ultimately, more effective data mining from taxonomic literature will significantly enhance taxonomic research and streamline the discovery and description of new species.
If you have used Floras or Faunas (hard copy or online) at any point in your work, research or leisure we'd love to hear from you. Perhaps you have used them in the past to try and identify species, to piece together the geographical distribution of particular taxa, to compile geographical or habitat based checklists of species, or to extract morphological or ecological traits from the descriptions. Even if your experience with them wasn't as successful as you'd hoped, we'd very much like to hear your views!
Producers of Floristic and Faunistic information face choices in the information that is prioritised, for example for digitisation and in the ways it is eventually presented to consumers. Often these important choices are based on an incomplete understanding of user needs, particularly of those users working outside of the taxonomic disciplines, but is also true to a lesser extent within the taxonomic community itself. The producers of information may be unaware exactly how their core biodiversity sources such as Floras and Faunas are being used, and which information is particularly valued. On the other hand, consumer groups who could benefit from primary information and associated services may be unaware of the existence of these resources, or else face other practical or technical barriers to their use.
Task 2.2 (within Workpackage 2) aims to identify the existing and the potential new consumers of Flora and Fauna information and services and to better understand their needs. A workshop is planned in May 2013 in Berlin. If you are interested in either filling out a short questionnaire or in attending the workshop please contact Soraya.Sierra [at] naturalis.nl
My PhD research programme is all about cladistic data congruence, compatibility, convergent evolution and phylogenetic tree-to-tree distance measures, while in parallel I work on a Panton Fellowship on data mining. The latter is about the mining of data direct from the literature and encouraging a culture of openness and data sharing. I shall be presenting this work at the pro-iBiosphere workshop in February 2013. My mentor for this project is Peter Murray-Rust, a Cambridge-based computational chemist and co-author of the Panton Principles for Open Data in Science. I'm fairly new to data mining techniques and machine learning methods, but am learning fast, and am certainly looking forward to meeting researchers at this event using similar techniques and methods for taxonomic data.
Specifically I'm looking to extract phylogenetic tree data direct from the figures of phylogenetic papers; including the exact relationships between taxa, branch lengths and support values. Unlike with some other data mining efforts that are entirely text-based, this requires some data extraction from non-textual sources. Some attempts have already been made to do this with programs like TreeThief, TreeRipper and TreeSnatcher but none of these are realistically and systematically applicable to tens of thousands of phylogenetic papers in their current state.
There is a huge wealth of phylogenetic data in the literature – I was co-author on a paper recently that shows that there are more than 66,000 separate papers containing novel empirically-generated phylogenetic trees in just the 21st century, and that less than 4% of these data are publicly available in a re-usable form. I, and many others, think this hugely valuable and repurposable data should be kept and made openly available for re-use, hence I'm trying to systematically salvage it from the literature.
Another novel aspect of our approach is that we're mining PDF's rather than publisher-provided XML or HTML. The latter do not contain the figures, just links to them, and thus they can only help us recover metadata on each phylogenetic tree. The PDF is often the only format in which it's all there and sometimes in clearly machine-interpretable format. Peter has been particularly vocal on his blog about the quality, or lack, of PDF files produced by some publishers on behalf of authors. Personally I feel that all supporting data for a paper should be made openly available, as the 'default' with exceptions to this rule only allowed with clear and explicit justification. I'm still surprised, and slightly disappointed, that this isn't yet the norm in scientific publishing – we certainly have the technology to do this. In 2013 I shall be submitting my PhD thesis; looking for further academic funding/employment and will assume the new role of Science Community Coordinator at the Open Knowledge Foundation – working to join together all the various open science labs around the world. I heartily look forward to meeting everyone at February’s pro-iBiosphere workshop.
Ross Mounce
PhD Student & Panton Fellow, University of Bath, United Kingdom
Journals not only disseminate information, they also provide a mechanism of quality control and certification for the results published. Species names and descriptions are the primary metrics in quantifying biodiversity, including communicating information about food and agriculture, ecologically important species, pests, and pathogens, and species of popular and conservation interest. For taxonomy, the close link between publication and research is even more crucial, as publications are the legal document validating the names of organisms.
Moving online increases accessibility to taxonomic information and ensures the long-term preservation through electronic archiving. However, this is a mission that requires public sector commitment and funding. Initiated under the umbrella of the European Distributed Institute for Taxonomy (EDIT), the European Journal of Taxonomy (EJT) is an example of institutions adapting to modern technologies so as to better fulfil their public mission.
Launched in September 2011, EJT is jointly published by a consortium of European natural History Institutes (NHI), namely those of Paris, London, Brussels, Meise, Tervuren, and Copenhagen, who have pooled their resources to publish an international, fully electronic, fast-track, peer-reviewed, non-for-profit and fully open access journal in descriptive taxonomy, covering subjects in zoology, entomology, botany (including mycology and algology), and palaeontology. There are no size limits to articles. EJT’s scope is global; authorship and geographical region of study are not exclusively European.
EJT supports the need to deposit type specimens in public collections (e.g. museums, herbaria). This policy anchors the title in a collection-based research environment.
The journal builds on the three main principles: high scientific quality, electronic and permanent Open Access archives; no financial costs to authors or readers.
The creation of this journal sends a strong political message to national and international funders of natural history research, showing the interest and capacity of NHIs in different countries to join forces and collectively claim a significant role in the organization of access to and dissemination of scientific information in their domain of research. By publishing their own joint journal, the institutions will be able to set conditions of access to the publicly funded research they perform.
Run by an editorial and a production team scattered throughout the NHIs that own and fund the title, EJT builds a European cross-institutional cooperation through light governance, enhancing coordination, establishing a cross-institutional strategy at the European level. EJT is designed to encourage and promote networking between publishing staff in NHIs and the biodiversity production and user community.
By addressing the current barriers for e-publishing, EJT aims at helping institutional journals within NHIs to movetheir publications to the Web efficiently and thus spread their scientific results more broadly and increase their citability and accessibility.
EJT’s team particularly welcomes pro-iBiosphere project and shares its main objectives: to enhance interoperability through coordination and promote the adoption of technological standards which are the only way to facilitate the access, dissemination, and use of scholarly publications in the fields of environmental and natural history.
We warmly invite you to explore the possibilities of publishing in EJT today, just click to www.europeanjournaloftaxonomy.eu
Laurence Bénichou
Responsable d'éditions/Publications manager Publications scientifiques du Muséum & European Journal of Taxonomy
Wouldn't it be lovely, when writing a paper, or compiling data, for the computer to tell you if you have spelled the name correctly, or that that the name has been superseded because of some taxonomic activity. Wouldn't it be nice to read documents and databases, have older names replaced with current names of organisms brought up to date automatically. This is the kind of functionality that is being pursued through the Global Names project.
The project capitalizes on the almost universal use of the Linnaean system of Latin binomials to annotate most of our meaningful observations of life made over the past 250 years. Those names offer us a way of indexing and linking information about species; whether in the 500 million or so estimated pages of literature; the billions of specimens located in museums and herbaria or in the tens of thousands of web sites and databases. Unfortunately, we don't have a unique name for each species because species may be split or moved from one genus to another. Nor is every name unique because the codes of nomenclature allow for the same name to be used for a plant and an animal. Because of this, we have to build an infrastructure that is taxonomically intelligent.
The potential of delivering names as part of the mechanism of integrating data distributed across the internet led GBIF and the Encyclopedia of Life to set up a series of Nomina workshops to conceive of what a future names-based cyberinfrastructure might be like. Last year, the USA’s National Science Foundation funded a two year 'Global Names' project (globalnames.org). Our goal is to provide openly available infrastructural tools that will assist in management of information about biodiversity. We have been building databases of names, developing various names discovery and names matching tools - such as the Chrome Names Spotter plug-in or the name discovery tool at http://gnrd.globalnames.org/). New names parsing algorithms help to convert name strings into canonical forms, or to offer the mechanisms for searches and browsing based on dates, authors, or genera. Our code is openly available at GitHub. Numerically, the most significant challenge lies with incorrectly spelled names that create variant 'name-strings' that prevent information on the same species in different data bases do be joined together. The solution is 'reconciliation', a process that maps all alternative names for the same species to each other so that a search initiated with one name can lead to an action that calls on all names. We hold over 22 million name strings, and there are many more to come, especially as older texts are digitized and OCR'd. We have extended the Rees / Giddens fuzzy matching tools to map variant spellings against each other.
Working with the Biodiversity Heritage Library, we have built a new indexing tool that includes names recognition, names discovery, names parsing, and a validation service. In the future, as necessary internal databases get increasingly populated, we can offer more validation services that will, for example, help biologists who are compiling research data or are writing papers to ensure that their names are spelled correctly, that they have the correct authority information, or that the name is the most current one for that taxon.
Our vision is of an open and very flexible cyberinfrastructure that all projects can contribute to and draw from so that we do not have to build multiple copies of databases. The result will be a more flexible and relevant suite of databases and services that will make it increasingly easier to discover and interconnect data. There remain many challenges, such as the capture of 250 years worth of synonymy information, full integration of vernacular names, and integration of the 'surrogates' for names that are increasingly flooding out of environmental surveys that rely solely on molecular techniques.
Reading: Patterson, D. J., Cooper, J., Kirk, P. M., Pyle, R.L. and Remsen D. P. 2010. Names are key to the big new biology. TREE 25: 686-691, doi:10.1016/j.tree.2010.09.004
David Patterson
dpatterson@mbl.edu
There are several important requirements for providing information on alien species. The information needs to be up-to-date, reliable and comprehensive, while its publication needs to be available to a wide range of stakeholders, clearly written and well illustrated. Traditional print media cannot fulfill all of these roles, but internet publication can.
The Manual of the Alien Plants of Belgium is a comprehensive guide to the plants that have been introduced to, and grow wild in Belgium. This includes a wild variety of plants, ranging from vigorous invasive species to rare casuals. Some are important agricultural weeds, some are escaped garden plants. In an era of environmental change, alien species occupy newly created niches. Halophytes spread along salted roads, new weeds grow amongst new crops; mountain plants find homes in the walls of cities and aquarium plants escape into ponds and rivers. Many of these plants cause real environmental and economic damage.
The Manual has been created using the Scratchpad system. The ability to add, correct, move and delete content at a moment’s notice means that the Manual can be reactive to new discoveries, to changes in taxonomy and to improve upon illustrations as they become available. It also allows the publication of a wide variety of other material including related publications, diagnostic keys and images of herbarium specimens.
The pro-iBiosphere project aims to promote IT publishing tools such as Scratchpads to a wider range of taxonomists. Tools such as these will make authoritative taxonomic, conservation and distributional information available to a much larger audience. These tools will allow much more efficient data reuse and the remobilization of legacy literature. Belgium might be a small country and alien plants might seem like an obscure subject. However, Belgium has a dense population; is an important transport hub; is intensive in agriculture and has hosts many industries, so despite its size it is highly susceptible to colonization by non-native species and dissemination of information on Belgium’s colonists has relevance far beyond its borders.
Quentin Groom & Filip Verloove
National Botanic Garden of Belgium
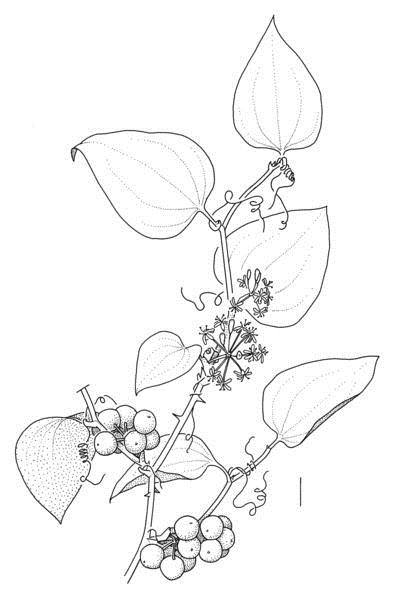 Smilax rotundifolia L., a rare alien in Belgium, which was presumably introduced as a garden plant. It is originally from the eastern United States. Drawn by Sven Bellanger
Smilax rotundifolia L., a rare alien in Belgium, which was presumably introduced as a garden plant. It is originally from the eastern United States. Drawn by Sven Bellanger
On 1st December 2012, 30 research institutions from 15 European countries, Brazil, Israel and the Philippines, and more than 30 associated partners started EU BON - "Building the European Biodiversity Observation Network". This €9 million, EU-funded research project aims to advance biodiversity knowledge by building a European gateway for biodiversity information, which will integrate a wide range of biodiversity data – both from on ground observations to remote sensing datasets – and make it accessible for scientists, policy makers, and the public.
The project plans to advance the technological platform for GEO BON (Group on Earth Observations Biodiversity Observation Network) to improve the assessment, analysis, visualisation and publishing of biodiversity information, and to enable better linkages between biodiversity and environmental data. EU BON will ensure a timely provision of integrated biodiversity information needed to meet the global change challenges and to contribute for next generation environmental data management at national and regional levels.
EU BON will deliver several important products, including a European integrated biodiversity portal, a roadmap for EU citizen sciences gateway for biodiversity data, an open data publishing and dissemination framework and toolkit, a policy paper on strategies for data mobilisation and use in conservation, a prototype of integrated, scalable, global biodiversity monitoring schemes, strategies for EU-integrated national and regional future biodiversity information infrastructures, and a sustainability plan for regional and global biodiversity information network.
The cooperation for data integration between biodiversity monitoring, ecological research, remote sensing and information users will result in proposing a set of best-practice recommendations and novel approaches with applicability under various environmental and societal conditions. A key task of EU BON is to harmonise future biodiversity monitoring and assessments and to engage wider society groups, such as citizen scientists and other communities of practise. Although focussing primarily on European biodiversity and collaborating with major EU initiatives (e.g. LifeWatch and others), EU BON will also collaborate closely with worldwide efforts such as GEO BON, GBIF, the Convention on Biological Diversity (CBD), the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services (IPBES) and others. EU BON will be a valuable European contribution to the Global Earth Observation System of Systems (GEOSS), and be built on the GEO principles of open data sharing.
The kick-off meeting of EU BON will take place on 13-15 February 2013 at the Museum für Naturkunde – MfN in Berlin, Germany and will be preceded by a symposium "Nature and governance: biodiversity data, science and policy interface" on 11-12 February 2013.
Additional information
EU BON (2012–2017) stands for "Building the European Biodiversity Observation Network" and is an European research project, financed by the 7th EU framework programme for research and development (FP7). EU BON seeks ways to better integrate biodiversity information and implement into policy and decision-making of biodiversity monitoring and management in the EU.
GEO BON stands for "Group on Earth Observations Biodiversity Observation Network". It coordinates activities relating to the Societal Benefit Area (SBA) on Biodiversity of the Global Earth Observation System of Systems (GEOSS). Some 100 governmental, inter-governmental and non-governmental organisations are collaborating through GEO BON to organise and improve terrestrial, freshwater and marine biodiversity observations globally and make their biodiversity data, information and forecasts more readily accessible to policymakers, managers, experts and other users. Moreover, GEO BON has been recognized by the Parties to the Convention on Biological Diversity. More information at: http://www.earthobservations.org/geobon.shtml.
GEOSS stands for Global Earth Observation System of Systems, built by the Group on Earth Observations (GEO). GEO is constructing GEOSS on the basis of a 10-Year Implementation Plan for the period 2005 to 2015. The Plan defines a vision statement for GEOSS, its purpose and scope, expected benefits, and the nine "Societal Benefit Areas" of disasters, health, energy, climate, water, weather, ecosystems, agriculture and biodiversity.
Link to the original press release.

Herbarium Bogoriense, Research Center for Biology-LIPI, is honored to be hosting the 9th International Flora Malesiana Symposium that will be held in Bogor on August 27-31, 2013. We would like to inform you that the website for the Symposium has been uploaded. Please visit http://www.fm9.biologi.lipi.go.id/ for more details about the symposium and program.
PDR 2012, 34(6) 623-630 DOI: 10.3724/SP.J.1143.2012.12139
PENG Hui-Fu, WANG Yu-Hua
In order to meet the broad requirements of the public and state departments to recognize and distinguish plant species, a plan for the next-generation Flora was proposed, which is internet, DNA-sequence- and information-technology- based, i.e., iFlora. In this paper, we suggest and present a research framework of "Scientific-Cloud for iFlora", which is a combination of Resource-Cloud (Resource and Service layer), Intelligent Re-organization System, Mobile-Clients (Application layer) and Key-Users. (1) Resource-Cloud: a collaborative working platform in the cloud site. This platform is used to integrate all relevant information resources, including DNA barcoding reference library, plant biodiversity information, and analysis models. (2) Intelligent Re-organization System of Virtualization: a system to retrieve, re-organize and present iFlora-records of a given plant species automatically and intelligently. (3) Mobile-Clients: software applications of mainstream mobile platforms, such as iOS, android and Windows phone. Constructed by these three parts, the framework will not only service for the key users with co-operating platform but also providing web service for the general public. The construction of this project will gradually combine botany and informatics. We are aiming to construct a sharing, easily accessible, and seamless connected Scientific-Cloud, with which numerous scientists co-operate together for iFlora. In addition, the Scientific-Cloud will integrate cognitive abilities of the machine, network, and taxonomists. It will provide an intelligent public service to perceive the plant world in a new dimension.
Source: A Framework of Scientific-Cloud for iFlora.
PLoS ONE (2012) 7(12): e50943. doi:10.1371/journal.pone.0050943
Duin D, King D, van den Besselaar P
Research evaluation should take into account the intended scholarly and non-scholarly audiences of the research output. This holds too for research infrastructures, which often aim at serving a large variety of audiences. With research and research infrastructures moving to the web, new possibilities are emerging for evaluation metrics. This paper proposes a feasible indicator for measuring the scope of audiences who use web-based e-infrastructures, as well as the frequency of use. In order to apply this indicator, a method is needed for classifying visitors to e-infrastructures into relevant user categories. The paper proposes such a method, based on an inductive logic program and a Bayesian classifier. The method is tested, showing that the visitors are efficiently classified with 90% accuracy into the selected categories. Consequently, the method can be used to evaluate the use of the e-infrastructure within and outside academia.
Source: Identifying Audiences of E-Infrastructures - Tools for Measuring Impact (PloS)
PLoS ONE 7(5): e36881. doi:10.1371/journal.pone.0036881
Fontaine B, van Achterberg K, Alonso-Zarazaga MA, Araujo R, Asche M, et al.
The number of described species on the planet is about 1.9 million, with ca. 17,000 new species described annually, mostly from the tropics. However, taxonomy is usually described as a science in crisis, lacking manpower and funding, a politically acknowledged problem known as the Taxonomic Impediment. Using data from the Fauna Europaea database and the Zoological Record, we show that contrary to general belief, developed and heavily-studied parts of the world are important reservoirs of unknown species. In Europe, new species of multicellular terrestrial and freshwater animals are being discovered and named at an unprecedented rate: since the 1950s, more than 770 new species are on average described each year from Europe, which add to the 125,000 terrestrial and freshwater multicellular species already known in this region. There is no sign of having reached a plateau that would allow for the assessment of the magnitude of European biodiversity. More remarkably, 1over 60% of these new species are described by non-professional taxonomists. Amateurs are recognized as an essential part of the workforce in ecology and astronomy, but the magnitude of non-professional taxonomist contributions to alpha-taxonomy has not been fully realized until now. Our results stress the importance of developing a system that better supports and guides this formidable workforce, as we seek to overcome the Taxonomic Impediment and speed up the process of describing the planetary biodiversity before it is too late. (full article text)
BMC Biology 2012, 10:87 doi: 10.1186/1741-7007-10-87
Miller J, Dikow T, Agosti D, Sautter G, Catapano T, Penev L, Zhang ZQ, Pentcheff D, Pyle R, Blum S, Parr C, Freeland C, Garnett T, Ford LS, Muller B, Smith L, Strader G, Georgiev T, Bénichou L.
Taxonomy is a fundamental science that provides the scaffolding for biology. But the true value of taxonomic data remains unrealized because basic biodiversity information remains fragmented and unevenly accessible. Taxonomy helps us recognize species and map their distributions by generating text descriptions, images, and records of when and where they have been observed. Current rates of species extinction, habitat loss, and climate change mean that taxonomy has never been more relevant. Biodiversity is one of the most information-rich fields of human knowledge , but advances in basic cybertaxonomic infrastructure have only recently provided the tools to organize biodiversity information in ways that respond to a wide range of user groups, including ecologists, land managers, and interested citizens, not to mention the benefits of readily accessible information to the global taxonomic community. The call to revitalize taxonomy by embracing the internet has been sounded for more than a decade. The time is ripe to significantly increase the volume of taxonomic information freely available online. But simply posting information online will not automatically reinvigorate taxonomy. There are myriad online sites dedicated to particular taxa or projects. These are useful to users interested in questions within the site's domains. But the greater potential lies in mechanisms for aggregating primary source data in ways that allow users to filter and recombine data easily and flexibly for whatever purposes they imagine. full article
The FP7 eInfrastructure project proiBiosphere targeting the preparation of the European Open Biodiversity Knowledge Management System was launched in Leiden in September 2012
The first article acknowledging pro-iBiosphere published: Penev L, Catapano T, Agosti D, et al. Implementation of TaxPub, an NLM DTD extension for domain-specific markup in taxonomy, from the experience of a biodiversity publisher. In: Journal Article Tag Suite Conference (JATS-Con) Proceedings 2012 [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2012. Available from: http://www.ncbi.nlm.nih.gov/books/NBK100351/. It is a result of WP 3 (survey and analysis of the status quo) in combination with an earlier review of XML-schemas in taxonomy.





 pro-iBiosphere wiki platform
pro-iBiosphere wiki platform
 RSS news
RSS news









