Lyubomir Penev, Gregor Hagedorn, Soraya Sierra
The latest Biodiversity Information Standards (TDWG) conference took place in Florence, Italy on October 28 – November 1, 2013. The pro-iBiosphere project was well represented in the conference with 8 participants. Consortium members contributed especially to the discussions about:
-
the future use of identifiers
-
re-establishing the Structured Descriptive Data standard in Semantic Web compatible RDF Format
-
future developments in DarwinCore
Presentations given by consortium members during the meeting are available here.
In order to make fundamental biodiversity data digital, open and re-usable, the pro-iBiosphere project has the vision of implementing an Open Knowledge Biodiversity System (OBKMS). The strong general current towards semantic technologies observed at the meeting, confirmed the need for this vision.
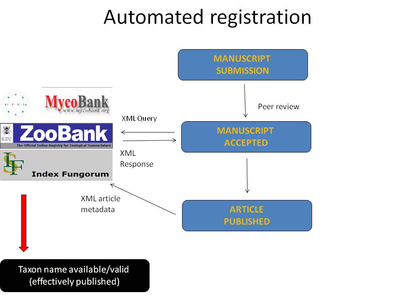
The conference is also to be remembered with the launch of the common automated registration model for higher plants, fungi and animals (one of the four pilots being conducted by the pro-iBiosphere project). Currently registration of new taxa is being conducted "by hand" by the authors, journals or registries. Pensoft journals were first to develop and offer this new service, in a collaboration with the International Plant name Index (IPNI), ZooBank and Index Fungorum. Detailed information on the automated registration pilot is available here.
Preceding the TDWG conference, Pensoft and ZooBank organised a hackathon in Sofia (Bulgaria). The hackathon resulted in several, real-time tests of the pilot, based on the TaxPub XML schema.
After the hackathon, the teams of IPNI, Index Fungorum, ZooBank, Pensoft and Plazi came together at a brief workshop to discuss the potential of the Taxonomic Concept Schema (TCS) as a possible basic model that may be used in the future for registration of taxa in the three organismic domains.
The consortium strives towards having a more open pilot registration system available by the end of the pro-iBiosphere project. At present, IPNI is having discussions with other key partners such as the IAPT committee on registration on how to open up the automated registration model.
Please mark your calendar for the pro-iBiosphere final event!
The final meeting will take place on the 10-12th of June 2014 in Belgium.
The National Botanic Garden of Belgium (NBGB) will be host to this meeting and has opened the doors of the Bouchout Castle for this event.
During the meeting you will have the opportunity to learn about outcomes of the project, be informed about relevant topics presented by invited speakers, follow training on state-of-the art tools, and do networking with colleagues.
We will keep you updated as our preparations develop.
In case of further questions please do not hesitate to contact us at pro-ibiosphere@sigma-orionis.com.
We look forward to seeing you in June!

The final EUBrazilOpenBio newsletter for November 2013 is now available. The newsletter presents the final press release showcasing the main results of 2 year collaborative work, namely: the innovative, web-based working environment designed to serve biodiversity scenarios; the new version of the Catalogue of Life cross-mapping tool developed in the i4Life project; the provision of the Ecological Niche Modelling tool as a service through the openModeller extended web service, and its application in collaboration with BioVeL; the EUBrazilOpenBio Joint Action Plan.
This newsletter highlights:
- EUBrazilOpenBio Joint Action Plan - drawing on policy strategies, analysing current progress in contributing to international targets and defining actions for future collaborative research. It defines common actions with the aim of contributing to relevant Aichi Targets in the years ahead.
- EGI federatec use case on ecology - Over the last two years, BioVeL and EUBrazilOpenBio have joined forces to make openModeller ready for cloud deployment. Work within the EGI Federated Cloud Task Force has led to considerable success in enabling the openModeller service on the EGI Federated Cloud.
- EUBrazilOpenBio results - EUBrazilOpenBio Technical developments, training materials and sessions, publications and papers, media spotlights and policy results all collected in one page.
- A vision from the Experts - "The Crossmapper itself is a great tool, and an ideal way to identify errors and updates". Dr Christina Flann is one of the experts providing their vision on EUBrazilOpenBio story.
You can find an online version of the final EUBrazilOpenBio newsletter here.

Prof. Robert E. Kenward [1]
A project to design a Transactional Environmental Support System (www.tess-project.eu) preceded pro-iBiosphere during 2008-1011. TESS, of which partners were mostly linked to the Sustainable Use and Livelihoods Group of IUCN (www.iucn.org/SULi), recognised the need for information on species and habitats to inform policy, but also at local level for management of land and sustainable use. TESS surveyed information needs at EU level, national governments, local communities at lowest government administrative level and stakeholders. The surveys not only showed biodiversity information requirements in detail, but also found a "decision density" five magnitudes higher for local managers of land and species (in farms, forests, fisheries, hunting areas and nature reserves) than for statutory environmental assessments (SEA+EIA).
To make decisions that are good for nature, there is scope for internet exchange (transaction) of science-based decision support (from high level) for the detailed data needed to make those local decisions (but also needed for policy-making at higher level). For effective exchange, data needs to be open access, standardised in format and highly detailed (accurate and at high resolution), i.e. aspirations of pro-iBiosphere. Case studies showed that local communities were highly capable of collecting appropriate data, but that thousands of models for science-based decision support included only 10 suitable for local use, at most in two languages!
This information was of special relevance for one of the pro-iBiosphere workshops in October 2013, Workshop on User Engagement and Benefits, showing in particular the importance of delivering Faunas, Floras and Mycotas for local people to record biodiversity. Identifying and reporting species needs to be made simple and multilingual for users other than experts, which could involve smart-phone apps for image-recognition and database-linking.
As part of its surveys, TESS established a portal (www.naturalliance.eu) to signpost relevant information and encourage collection by local interests. Naturalliance is advised by major international NGOs at European and Global level, is now in 23 languages and, to encourage sustainable use of land and biodiversity, and is working with national governments on a new system to encourage systematic local participation in data exchange. Experience with funding these systems, which could signpost a one-stop-shop for identification/reporting apps, or even host the downloading, was shared during the meeting on 10 October 2013 to evaluate business models currently in use by partners and relevant non-partners.
[1] Vice-chair (Europe) of IUCN Sustainable Use and Livelihoods Specialist Group
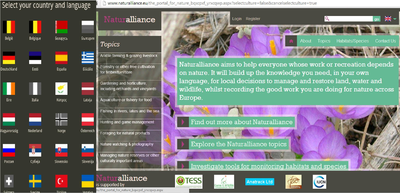 The Naturalliance Portal developed by IUCN-SULi during the TESS project.
The Naturalliance Portal developed by IUCN-SULi during the TESS project.
Hanna Koivula[1]
The Finnish Museum of Natural History is a focal point for GBIF Finland (FinBIF). For the past few years we have been constructing a national research infrastructure to serve not only data needs of biodiversity researchers, but also environmental services. The aim is to fulfill many different data requirements for various kinds of surveys and assessments concerning biodiversity. Most of our data are openly available to anyone, but technically many data users have not been able to use our web services for retrieving the data they need. The main obstacle is that the services are very new and the awareness about exchange protocols and standards of the data is poor. This is why most of the data shared by FinBIF are still extracted manually upon request. Hence, one (still unreached) goal is to name and establish best practices for sharing and maintaining biodiversity data. This is done by applying selected parts, especially for logical architecture and communication, from Enterprise architecture. The method enables us to capture essential information of our research infrastructure under construction.
Building a research infrastructure in an extremely heterogeneous "funding environment" has set some serious impediments along the way. On the other hand, it has allowed us to play with quite adventurous architectural structures and innovative pilot cases. One of these new approaches is the use of semantic web features for making data storage more flexible and data themselves better discoverable.
Participating in the pro-iBiosphere workshop on "How to improve technical cooperation and interoperability at the e-infrastructure level" that took place on the 8th of October 2013, in Berlin, gave me a good insight into existing European open biodiversity data and related services. Hands-on experience of listing our web services to the Biodiversity Catalogue triggered useful conversations on how to improve the interoperability and discoverability of the services, and gave participants take-home ideas. In the break-out discussion group we assessed the present set of metadata elements used in the Biodiversity Catalogue for describing individual services. Based on that evaluation, we made recommendations for additional elements. I also obtained many good ideas for further developing the FinBIF services.
[1] Finnish Museum of Natural History - IT Specialist GBIF Node Manager for Finland, Email: hanna.koivula@helsinki.fi
Sylvia Mota de Oliveira[1]
The three countries forming the Guianas - Guyana, Suriname and French Guiana - have more than 80% of their political territories covered by pristine Amazon forest. The Flora of the Guianas book series provides taxonomic treatments and associated data on plant species occurring in the region. At present, the published fascicles cover around 25% of the total of species recorded in the Guianas, with a total of 2294 species treated only in series A: Phanerogams and around 1000 species in other series, covering Ferns and Fern allies, Bryophytes and Fungi. Beyond taxonomy, the Flora is a rich and well curated source of primary biodiversity data. To efficiently use this source and build it further, we want to move towards an online platform, where:
- Data extracted from other literature sources (Flora of Suriname, for instance) can be incorporated in a database and used as a basis for upcoming taxa treatments, increasing the rate of species descriptions;
- Data gathered in the Flora fascicles can be disclosed and made available in a structured and searchable manner, openly accessible to end-users;
- Interaction between Flora contributors, specialists and citizen scientists is promoted;
- Links between elements of the Flora and external content, such as collection and molecular databases, can be easily enabled;
- Updates of the taxonomic data can guarantee the quality and usability of the Flora in several other research fields.
To achieve these goals, the transference of the data from the printed copies of the Flora of the Guianas to such an online platform requires mark-up of text. The new workflow is being tested in a Pilot study within the pro-iBiosphere project, which will hopefully support the planning of a longer-term strategy for the complete Flora.
The first results of the pilot were presented and discussed during the pro-iBiosphere workshop on "How to promote and foster the development & adoption of common mark-up standards & interoperability between schemas" that took place on the 8th of October 2013, in Berlin. The workshop was very instructive and useful for the future strategy of the Flora of the Guianas. In this workshop, participants shared their experiences, and discussed together the most important issues/challenges that they have encountered while working on text mark-up. The heterogeneity of the group added to the discussion because participants were confronted with different levels of: 1) experience with mark-up tools; 2) data granularity; 3) consistency in the format of the content to be marked-up. Relevant topics discussed included the scaling of the mark-up activities using different tools, the costs and possible incentives for such an activity, the desired level of granularity of the mark-up, and best practices to be adopted by taxonomists and bioinformaticians.
[1] Editor of Flora of the. Guianas, Naturalis Biodiversity Center, the Netherlands. Contact: Sylvia.MotaDeOliveira@naturalis.nl
Zookeys, the first zoological journal to introduce automatic registration in ZooBank
The latest issue of ZooKeys - no. 346 - has been automatically registered in ZooBank on its day of publication last Friday. This marks the successful deployment of an automated registration-to-publication pipeline for taxonomic names for animals. The innovative workflow was jointly funded by the EU FP7 funded project pro-iBiosphere and a U.S. National Science Foundation project to develop the Global Names Architecture (DBI-1062441).
The process of post-publication recording and indexing of species names has a long tradition, in some cases dating as far back as the middle of 19th century. But now in the 21st century with the advance of modern technologies and the opportunity to publish taxonomic novelties online, the process of post-publication recording brought into focus the concept of automated pre-publication registration.
Why is this important? The proportion of 'turbo-taxonomic' papers describing hundreds of new species increases. Registration of hundreds of new species is an issue, however it is even more important that the final publication data of the pre-registered names are reported back to ZooBank on the day of publication.
Launched as an open access peer reviewed journal in 2008, to coincide and adopt from inception the International Code of Zoological Nomenclature changes for electronic publications, ZooKeys was the first journal to provide a mandatory in-house registration in ZooBank. Since 2008, it has contributed about one third of all names currently registered in ZooBank. With the adoption of the automated ZooBank registration, ZooKeys continues its mission to set novel trends in biodiversity publishing.
Implementation of automated workflows and invention of XML-based tools will facilitate the process of publication and dissemination of biodiversity information. It will pave the way for unification and streamlining the registration process, even more to building the next-generation e-infrastructure for a common global taxon names registry. Within the pro-iBiosphere project and in cooperation with Plazi that have created the TaxPub XML schema, an automated registration workflow for plants has already been established between the International Plant Names Index (IPNI) and the PhytoKeys journal, to be applied soon also for fungi between Index Fungorum and the journal MycoKeys.
Taxonomic descriptions, introduced by Linnaeus in 1735, are designed to allow scientists to tell one species from another. Now there is a new futuristic method for describing new species that goes far beyond the tradition. The new approach combines several techniques, including next generation molecular methods, barcoding, and novel computing and imaging technologies, that will test the model for big data collection, storage and management in biology. The study has just been published in the Biodiversity Data Journal.
While 13,494 new animal species were discovered by taxonomists in 2012, animal diversity on the planet continues to decline with unprecedented speed. Concerned with the rapid disappearance rates scientists have been forced towards a so called 'turbo taxonomy' approach, where rapid species description is needed to manage conservation.
While acknowledging the necessity of fast descriptions, the authors of the new study present the other 'extreme' for taxonomic description: "a new species of the future". An international team of scientists from Bulgaria, Croatia, China, UK, Denmark, France, Italy, Greece and Germany illustrated a holistic approach to the description of the new cave dwelling centipede species Eupolybothrus cavernicolus, recently discovered in a remote karst region of Croatia. The project was a collaboration between GigaScience, China National GeneBank, BGI-Shenzhen and Pensoft Publishers.
Eupolybothrus cavernicolus has become the first eukaryotic species for which, in addition to the traditional morphological description, scientists have provided a transcriptomic profile, DNA barcoding data, detailed anatomical X-ray microtomography (micro-CT), and a movie of the living specimen to document important traits of its behaviour. By employing micro-CT scanning in a new species, for the first time a high-resolution morphological and anatomical dataset is created - the 'cybertype' giving everyone virtual access to the specimen.
This, most data-rich species description, represents also the first biodiversity project that joins the ISA (Investigation-Study-Assay) Commons, that is an approach created by the genomic and molecular biology communities to store and describe different data types collected in the course of a multidisciplinary study.
"Communicating the results of next generation sequencing effectively requires the next generation of data publishing" says Prof. Lyubomir Penev, Managing director of Pensoft Publishers. "It is not sufficient just to collect 'big' data. The real challenge comes at the point when data should be managed, stored, handled, peer-reviewed, published and distributed in a way that allows for re-use in the coming big data world", concluded Prof. Penev.
"Next generation sequencing is moving beyond piecing together a species genetic blueprint to areas such as biodiversity research, with mass collections of species in "metabarcoding" surveys bringing genomics, monitoring of ecosystems and species-discovery closer together. This example attempts to integrate data from these different sources, and through curation in BGI and GigaScience's GigaDB database to make it interoperable and much more usable," says Dr Scott Edmunds from BGI and Executive Editor of GigaScience.
Additional information:
Pensoft and the Natural History Museum London have received financial support by the EU FP7 projects ViBRANT and pro-iBiosphere. The China National GeneBank (CNGB) and GigaScience teams have received support from the BGI. The DNA barcodes were obtained through the International Barcode of Life Project supported by grants from NSERC and from the government of Canada through Genome Canada and the Ontario Genomics Institute.
Original Sources:
Stoev P, Komerički A, Akkari N, Shanlin Liu, Xin Zhou, Weigand AM, Hostens J, Hunter CI, Edmunds SC, Porco D, Zapparoli M, Georgiev T, Mietchen D, Roberts D, Faulwetter S, Smith V, Penev L (2013) Eupolybothrus cavernicolus Komerički & Stoev sp. n. (Chilopoda: Lithobiomorpha: Lithobiidae): the first eukaryotic species description combining transcriptomic, DNA barcoding and micro-CT imaging data. Biodiversity Data Journal 1: e1013. DOI: 10.3897/BDJ.1.e1013
Edmunds SC, Hunter CI, Smith V, Stoev P, Penev L (2013) Biodiversity research in the "big data" era: GigaScience and Pensoft work together to publish the most data-rich species description. GigaScience 2:14 doi:10.1186/2047-217X-2-14
Watch the 3D cybertype video: http://www.youtube.com/watch?v=vqPuwKG8hE4&feature=em-upload_owner

The Biodiversity Data Journal goes beyond the basics of the Gold Open Access
There are two main modes of open access publishing – Green Open Access, where the author has the right to provide free access to the article outside the publisher's web site in a repository or on his/her own website, and Gold Open Access, where articles are available for free download directly from the publisher on the day of publication.
Opening of content and data, however does not necessarily mean "easy to discover and re-use". The Biodiversity Data Journal proposed the term "Advanced Open Access" to describe an integrated, narrative (text) and data publishing model where the main goal is to make content "re-usable" and "interoperable" for both humans and computers.
To publish effectively in open access, it is not sufficient simply to provide PDF or HTML files online. It is crucial to put these under a reuse-friendly license and to implement technologies that allow machine-readable content and data to be harvested and collated into a big data pool.
The Advanced Open Access means:
- Free to read
- Free to re-use, revise, remix, redistribute
- Easy to discover and harvest
- Content automatically summarised by aggregators
- Data and narrative integrated to the widest extent possible
- Human- and computer-readable formats
- Community-based, pre- and post-publication peer-review
- Community ownership of data
- Free to publish or at low cost affordable by all
BDJ shortens the distance between "narrative" (text) and "data" publishing. Many data types, such as species occurrences, checklists, measurements and others, are converted into text from spreadsheets for better readability by humans. Conversely, text from an article can be downloaded as structured data or harvested by computers for further analysis.
"Open access is definitely one of the greatest steps in scientific communication comparable to the invention of the printing technology or the peer-review system. Great but not sufficient!" said Prof. Lyubomir Penev, founder of Pensoft Publishers and the Biodiversity Data Journal. "We need to switch the focus already from making content 'available for free download' to being discoverable and extractable. Such re-usability multiplies society's investment in science".
Additional information:
The Biodiversity Data Journal is designed by Pensoft Publishers and was funded in part by the European Union's Seventh Framework Program (FP7) project ViBRANT.
Source: Smith V, Georgiev T, Stoev P, Biserkov J, Miller J, Livermore L, Baker E, Mietchen D, Couvreur T, Mueller G, Dikow T, Helgen K, Frank J, Agosti D, Roberts D, Penev L (2013) Beyond dead trees: integrating the scientific process in the Biodiversity Data Journal. Biodiversity Data Journal 1: e995. DOI: 10.3897/BDJ.1.e995
A correspondence item, published today, 10 Oct 2013, in Nature focuses on the upcoming calls for Horizon 2020 research funding. The European Commission has said that it would prefer bids from open, collaborative consortia rather than the competitive bids seen in previous funding programmes. The authors call for an effort to forge interdisciplinary links in biodiversity research, and ask readers to contribute to discussions on project ideas.
For more information read the full correspondence item in Nature: http://www.nature.com/nature/journal/v502/n7470/full/502171d.html
Europe’s most visible forum for ICT research and innovation, ICT2013, will take place from 6-8 November 2013 in Vilnius, Lithuania.
The ICT 2013 event will be the first opportunity to learn the details of research funding for ICT-related projects under Horizon 2020, the EU’s new research program for 2014-2020. ICT2013 will also offer participants opportunities to showcase their most advanced research, ICT products and most innovative creations and to meet delegates with common or similar topical interests with whom they could collaborate in the future.
The pro-iBiosphere partnership will be represented at the ICT2013 event with a networking session and an exhibition booth, co-organised with other three EC-funded projects: the European Grid Infrastructure (EGI), ei4Africa and CHAIN-REDS.
The networking session on ‘What does the future hold for e-Science and Big Data?’ is taking place on Nov. 6, 2013 (16:00-16:45). This session will bring together researchers, data owners and service providers (including SMEs) to explore the future for e-science and how to deliver open access to data through Horizon2020. Participation will lead to better awareness of e-infrastructures and their potential for universal access to big data and a closer understanding of how they solve scientific challenges, including supporting science’s ‘long tail’. More…
The exhibition booth ‘e-Infrastructures at work and the future of research’ will showcase demonstrations from the four EC-funded projects running the booth. Members of the four projects will be on hand to fully engage with visitors and to present several demonstrators. There will also be supporting information and materials such as posters and leaflets.
We would be glad to meet you to discuss future collaboration opportunities in the fields of open access to data and linking litterature and data: if you would like to arrange a meeting with us, please contact us!
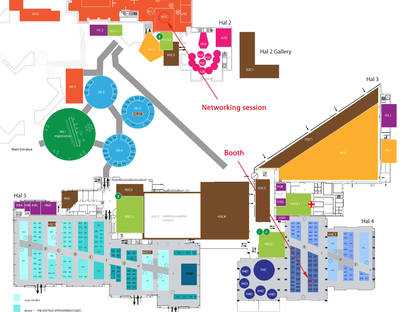
How to find us
The networking session will be organised in the Room H1C, located in the Hal 1. The exhibition booth (n. 4B24) will be located in the Hal 4: see map below.
Global Biodiversity Informatics Outlook sets out key steps to harness IT and open data to inform better decisions
Copenhagen, Denmark – A new initiative launched today (2 Oct) aims to coordinate global efforts and funding to deliver the best possible information about life on Earth, and our impacts upon it.
The Global Biodiversity Informatics Outlook sets out a framework to harness the immense power of information technology and an open data culture to gather unprecedented evidence about biodiversity and to inform better decisions.
The framework is outlined in a document and website entitled Delivering Biodiversity Knowledge in the Information Age, inviting policy makers, funders, researchers, informatics specialists, data holders and others to unite around four key focus areas where progress is needed.
The focus areas, each consisting of several specific components, are:
- Culture – promoting practices and infrastructure for sharing data, using common standards and persistent archives, backed up by strong policy incentives and a community of willing specialists;
- Data – addressing the need to transform all data about species, past and present, into usable and accessible digital formats; from historic collections and literature to citizen science observations, remote sensors and gene sequencing;
- Evidence – organizing and assessing data from all sources to provide clear, consistent views giving them context; including taxonomic organization, integrated occurrences in time and space, capturing information about species interactions, and improving data quality through collaborative curation; and
- Understanding – building models from recorded measurements and observations to support data-driven research and evidence-based planning, including predictive tools, better visualization and feedbacks to prioritize new data capture.
The document is being promoted through a number of upcoming events this month, including the Governing Board of the Global Biodiversity Information Facility and the Subsidiary Body on Scientific, Technical and Technological Advice of the Convention on Biological Diversity (CBD SBSTTA) where it forms part of the discussion on meeting global targets to end biodiversity loss.
The framework arose from the Global Biodiversity Informatics Conference which gathered around 100 experts in Copenhagen in July, 2012, to identify critical questions relating to biodiversity and tools needed answer them. Workshop leaders at that conference went on to draw up and author the current document.
The Global Biodiversity Informatics Outlook includes examples of projects and initiatives contributing to its objectives, and the accompanying website www.biodiversityinformatics.org invites feedback from others wishing to align their own activities to the framework.
A deck of slides for presentations about GBIO is available at http://www.slideshare.net/GBIF/global-biodiversity-informatics-outlook
The pro-iBiosphere project is organising 4 workshops to be held on October 8 -10, 2013 in Berlin:
- 8 Oct. - Workshop 1: How to improve technical cooperation and interoperability at the e-infrastructure level
- 8 Oct. - Workshop 2: How to promote and foster the development & adoption of common mark-up standards & interoperability between schemas
- 9 Oct. - Workshop 3: Workshop on user engagement and benefits
- 10 Oct. - Workshop 4: Towards sustainability towards service: Meeting to evaluate business models currently in use by partners and relevant non-partners
The workshops will take place at the Berlin Botanical Garden and Museum, Königin-Luise Straße 6-7, Berlin-Dahlem.
For complementary information on these events (concept and objectives, accommodation and transportation), please visit the dedicated project wiki page here or contact us: info@pro-ibiosphere.eu.
You are invited to follow and/or participate in the discussions on Twitter by using the hashtag #pibber !

The Biodiversity Data Journal (BDJ) and the associated Pensoft Writing Tool (PWT), launched on 16th of September 2013, offer several innovations - some of them unique - at every stage of the publishing process. The workflow allows for authoring, peer-review and dissemination to take place within the same online, collaborative platform.
Open access to content and data is quickly becoming the prevailing model in academic publishing, resulting in part from changes to policies of governments and funding agencies and in part from scientist's desire to get their work more widely read and used. Open access benefits scientists with greater dissemination and citation of their work, and provides society as a whole access to the latest research.
To publish effectively in open access, it is not sufficient simply to provide PDF files online. It is crucial to put them under a reuse-friendly license and to implement technologies that allow machine-readable content and data to be harvested by computers that can collate small scattered data into a big pool. Analyses and modelling of community-owned big data are the only way to confront environmental challenges to society, such as climate change, ecosystems destruction, biodiversity loss and others.
Manuscripts are not submitted to BDJ in the usual way, as word processor files, but are written in the online, collaborative Pensoft Writing Tool (PWT), that provides a set of pre-defined, but flexible article templates. Authors may work on a manuscript and invite external contributors, such as mentors, potential reviewers, linguistic and copy editors, and colleagues, who may read and comment on the text before submission. When a manuscript is completed, it is submitted to the journal with a simple click of a button. The tool also allows automated import of manuscripts from data management platforms, such as Scratchpads.
"This is the first workflow ever to support the full life cycle of a manuscript, from initial drafting through submission, community peer-review, publication and dissemination within a single, online, collaborative platform. By publishing papers in all branches of biodiversity science, including novel article types, such as data papers and software descriptions, BDJ becomes a gateway for either large or small data into the emerging world of "big data", said Prof. Lyubomir Penev, managing director and founder of Pensoft Publishers.
BDJ shortens the distance between "narrative (text)" and "data" publishing. Many data types, such as species occurrences, checklists, measurements and others, are converted into text from spreadsheets into a human-readable format. Conversely text from an article can be downloaded as structured data or harvested by computers for further use.
A novel community-based peer-review provides the opportunity for a large number of specialists in the field to review a manuscript. Authors may also opt for an entirely public peer-review process. Reviewers may opt to be anonymous or to disclose their names. Editors no longer need to check different reviewers' and author's versions of a manuscript because all versions can be consolidated into a single online document, again at the click of a button.
"The Biodiversity Data Journal is not just a journal, not even a data journal in the conventional sense. It is a completely novel workflow and infrastructure to mobilise, review, publish, store, disseminate, make interoperable, collate and re-use data through the act of scholarly publishing!" concluded Dr Vincent Smith from the Natural History Museum in London, the journal's Editor-in-Chief.
The platform has been designed by Pensoft Publishers and was funded in part by the European Union's Seventh Framework Program (FP7) project ViBRANT.
Original Source
Smith V, Georgiev T, Stoev P, Biserkov J, Miller J, Livermore L, Baker E, Mietchen D, Couvreur T, Mueller G, Dikow T, Helgen K, Frank J, Agosti D, Roberts D, Penev L (2013) Beyond dead trees: integrating the scientific process in the Biodiversity Data Journal. Biodiversity Data Journal 1: e995. DOI: 10.3897/BDJ.1.e995

After a successful first edition in 2011, the European Commission has launched the second edition of the EU Prize for Women Innovators to reward three women who have developed outstanding innovations and brought them to market.
The contest is open until 15 October 2013, 5:00 pm (Brussels time) to all women who have founded or co-founded their own company and who have at some point in their career benefited from the EU's research framework programmes or the Competitiveness and Innovation framework programme.
The first prize is EUR 100,000, second prize EUR 50,000 and the third prize EUR 25,000.
With this Prize, the European Commission aims to raise awareness about the contribution, potential and importance of Women researchers to entrepreneurship and to encourage women to exploit the commercial and business opportunities offered by their research projects and become entrepreneurs.
Compete and tell your story now to inspire other women to follow in your footsteps!
Applications can be submitted via the competition website until 15 October 2013 (5 p.m. Brussels time): www.ec.europa.eu/women-innovators
The traditional audience for books and scientific papers in which scientists report their findings has been the human reader. Now we can enhance publications by attaching to them many different kinds of digital objects (such as the sounds made by birds, maps that show where they occur, or images and videos) or by adding computer-readable sections and terms that allow computers to extract information for re-use. We refer to these enriched and marked-up documents as 'enhanced'.
While the technology is available, only a tiny proportion of scientific publications are enhanced. Without enhancement, the research that is reported in the biosystematic (= taxonomic) literature cannot participate quickly and easily in the big data world.
The EU e-Infrastructure coordination project "pro-iBiosphere", targeting the preparation of the European Open Biodiversity Knowledge Management System, makes thirteen recommendations to enhance the publication process in order to to make biodiversity data accessible, computable and re-usable. These recommendations are a plea for a major change in how we publish biodiversity research. If adopted, they have the potential to transform the role of scientific publications. The recommendations include the need to make all biosystematic literature "openly and freely accessible to the maximum extent possible and for it to be marked up with computer-legible terms from an open, platform-independent XML or similar language. Mark-up allows computers to understand what is in a document, and to extract content for use elsewhere. The complete list of recommendations is available on "The State and Quality of Biosystematics Documents and Survey Reports". Inputs are welcomed on proi-Biosphere's Google +, LinkedIn or Facebook.
The reports are based on two workshops in Leiden (February 2013) and Berlin (May 2013) organised by the project with 100 and 45 participants, respectively; a questionnaire answered by 60 persons including taxonomists and related professionals; literature review and conversations with taxonomists.
" I think pro-iBiosphere has the best sense of how to prepare for a future of biodiversity science. e-Flora, e-Faunas, e-Mycotas, e-Protistas, etc. are merely conduits for semantically ready biodiversity information to flow to end users"
"I really enjoyed the morning workshop sessions and think that the results from these morning sessions have great potential for helping shape how we best carry out biodiversity research in the future and reach our users most effectively"
said two participants of the May 2013 pro-iBiosphere workshop.
The report demonstrates that publishers and scientists are not using new technologies to their full potential. The process of publication has historically targeted scientific articles that are suited to human readership. Nowadays, articles can also be used as vehicles for data that can be extracted and then re-used by others. This process does not reduce the value of articles for people, but creates a richer and more valuable resource at a very small additional cost
The authors of the report recognise that the community must build an infrastructure that can capture and manage the rich supply of re-usable data, and make it available to other users, for example through the Linked Open Data Cloud. Only through this process, can the very rich corpus of data and its underlying sources in the published record be linked to the huge production of born-digital genomic data. With developments such as this, biologists will be better placed to rise to the grander challenges, such as understanding how the natural world will respond to a warming world.
By Donat Agosti, PLAZI
"Well, I suppose I'd better start finding names for things…" was the first thing said by the ill-fated sperm whale (Physeter macrocephalus Linnaeus, 1758) in Douglas Adam's Hitchhikers Guide to the Galaxy. Such is the richness of life that, unlike that whale, mankind is still naming things. Without commonly accepted names there would be no way to communicate research about life and it is the profession of taxonomists to put names on organisms and describe the different forms of life on Earth.
The EU e-Infrastructure coordination project "pro-iBiosphere", targeting the preparation of the European Open Biodiversity Knowledge Management System, makes ten recommendations to increase the adoption of digital workflows in the biodiversity domain. The recommendations include a "focus on usability and interoperability of software, not just functionality; fostering openness with research data"; and aim at giving further stimuli to the engagement of taxonomists with digital technology. The complete list is available in the report "The use of e-tools among producers of taxonomic knowledge". The document is based on a workshop and training organised by the project with 100 participants, a questionnaire answered by 220 persons including taxonomists and related professionals, literature review and conversations with taxonomists.
One of the attendees at the pro-iBiosphere workshops said "pro-iBiosphere is helping to close the gap between technology and taxonomists". Thibaut DeMeulemeester, Postdoctoral researcher (Naturalis Biodiversity Center, Leiden, the Netherlands).
Taxonomy has traditionally been a paper-based occupation and due to the rules on naming priority it relies heavily on the vast corpus of literature that has accumulated since the works of Carl Linnaeus. Yet, other scientists, conservationists and resource managers are crying out for a more transparent, stable, available and linked system of names. This can only be achieved digitally, openly and on the internet.
The need for a digital system of taxonomy is higher than ever. Modern metagenomic methods have been developed that can extract DNA sequence from 'soup' containing hundreds of species, many of which that cannot be linked to known species and some of which may in fact be new species to science. This is an exciting new technique with the potential to teach us much about microbial ecology, evolution and diversity. Yet it presents a challenge to taxonomy. The potential for generating thousands of DNA sequence from hitherto unrecognized species means that our ability to connect the genetic and biological data with their names will be severely tested. Only a digital online system of taxonomy can hope to keep up with these exciting developments.
pro-iBiosphere aims at binding together the disparate disciplines of the life sciences and envisages the digital-taxonomist providing the names that glue it all together.
Your feedback and suggestions are welcomed on pro-iBiosphere's Google +, LinkedIn or Facebook.
By Quentin Groom, NBGB

The EU e-infrastructure coordination pro-iBiosphere project is preparing the ground for the pursuit of biological research in the digital age. In its "Draft policy for Open Access to data and information" scientists and lawyers recommend that hurdles posed by copyright and database protection should be removed by establishing exceptions for research in a new binding, Europe-wide regulation. This report opens a consultation process that will last until December 2013. Input is welcomed on pro-iBiosphere's Google+ , LinkedIn or Facebook.
At present, national provisions on copyright and database protection regarding exceptions and limitations for research purposes differ both in detail and substance. Scientists within the EU working with copyright protected works or with protected databases have to be aware that regulations may vary considerably from country to country. This can be a major stumbling block to international collaboration in science.
The document addresses legal issues that hamper an integrative system for managing biodiversity knowledge in Europe. It describes the importance for scientists to have access to documents and data in order to synthesize disparate information and to facilitate data mining (or similar research techniques). It explores some aspects of copyright and database protection that influence access to and re-use of biodiversity data and information and refers to exceptions and limitations of copyright or database protection provided for within the relevant EU Directives.
The scientists also suggest that publicly funded institutions should refrain from claiming intellectual property rights for biodiversity data and information published or made accessible by them. Re-use of biodiversity data and information for research purposes should be allowed without any form of authorization. The only claims that publicly funded institutions should make are to ensure users fully acknowledge the sources of information that they rely on.
The report concludes that the legal situation within the EU is unsatisfactory and, hence, the creation of a much-needed integrative system for biodiversity knowledge will be hampered by copyright or by database protection. The scientific community recommends that these hurdles should be removed by unifying the terms that relate to research needs in a binding, Europe-wide regulation.
The vision pro-iBiosphere is to prepare the ground for an integrating global system for the intelligent management of biodiversity knowledge (i-Biosphere). Such system will:
- Offer a robust service-oriented architecture for distributed taxon-level information
- Include a central registry of sources and services, with documentation, so that they can be discovered
- Provide open and free access to all names and taxonomic information from a single source to all persons who need biodiversity data, without legal barriers, copyright and database protection rights, nor requiring the consent of other individuals or institutions
- Facilitate the re-use of biodiversity data and information
- Be interoperable with closely related initiatives
- Be fully aware of user requirements so that it serves the community as a whole
- Have a solid long term sustainability plan to maintain the infrastructure and the services
In order to fulfil this vision, technical and semantic interoperability challenges need to be addressed; user requirements need to be known; sustainability plans need to be developed; and basic requirements like allowing open and free access to data and information and re-usability for legitimate purposes need to be in place. At present, these basic requirements are hampered by numerous factual, technical, economic, sociological and other factors as well as by putative or real legal barriers, in particular, copyright and database protection rights.
By Willi Egloff, PLAZI
Two of the biggest challenges of aggregating data and giving credit to the sources are duplication and ambiguity. The easiest way to resolve this is through unique persistent identifiers to objects, events and people. In pro-iBiosphere we have discussed at length identifiers for specimens (http://wiki.pro-ibiosphere.eu/wiki/Best_practices_for_stable_URIs). However; a solution already exists for people. ORCID is an open, non-profit, community-driven organization that provides a registry of unique and persistent researcher identifiers.
ORCID is a hub that connects researchers and their professional activities through the embedding of ORCID iDs in key research workflows, data systems and other identifier systems. ORCID is unique in its ability to reach across disciplines, research sectors and national boundaries and in its cooperation with other identifier systems. The ORCID iD is a key component that supports system interoperability.
- ORCID improves discoverability and reduces repetitive data entry.
- ORCID reduces activity reporting, eases staff burden, and simplifies the manuscript submission processes.
- ORCID iDs overcome name ambiguity, distinguishing researchers and ensuring work is correctly attributed.
- ORCID supports a more comprehensive understanding of global research, investment and impact.
Working in collaboration with others in the research community — universities, research organizations, funders, publishers and professional associations — the embedding of ORCID iDs is widespread and includes integration in researcher profile systems, manuscript submission processes, HR systems, grant applications, and in linkages with repositories and other researcher identifier systems. ORCID benefits the entire research community, and relies upon its organizational members for support.
Registration is free for researchers, and they control the privacy of their ORCID record. It only takes seconds to register — visit ORCID today.
Quentin Groom (http://orcid.org/0000-0002-0596-5376)
Rebecca Bryant (http://orcid.org/0000-0002-2753-3881)
 ORCID logo
ORCID logo
On the 2nd July 2013, I attended the EC Public consultation on open research data.
The EU has already a commitment to open publication of publicly funded research, but this was a consultation on the policy for the openness of data. The European Commission recognizes that openness of data is better for scientific advancement, it promotes innovation and it is also good for the citizen. It means that scientific research will be more verifiable and it will promote acceptance of research.
There were five topics for consultation:-
1. What types of data should be open and how do you define what research data is?
2. What restrictions should be placed on openness and when?
3. Where should open data be stored and made accessible?
4. How can a culture of openness be promoted?
5. How can issues related to re-use, such as citation, be addressed?
There were many perspectives at the meeting, from industry, medical research, particle physics, publishers, research funders, librarians, etc. However, from a Biodiversity perspective I came away with several points that I thought worth sharing.
1. Biodiversity researchers are much better placed than many other fields to move towards openness. We are not often restricted by issues of personal privacy and commercially sensitive data.
2. Being open with data will cost money. It is still not easy for the majority of biodiversity scientists to get their data in a format that they can deposit in a central repository. This will require investment in software and training.
3. Well maintained central repositories are essential for the storage, dissemination and citation of open data.
4. Unique identifiers of people (e.g. ORCID), publications (e.g. DOI) and data are essential for a culture of openness.
5. Data management plans are likely to be required in Horizon 2020 proposals, either in the proposal or as a first deliverable. Data management plans are now becoming essential at national and institutional level.
We have a long way to go in the promotion of openness in the Biodiversity community. Other scientific communities are well ahead of us. Yet there are good reasons to be optimistic, particularly with the progress of open publication. A change to a culture of sharing will develop if issues of citability of data can be resolved and if institutions give recognition to researchers for being open with their data.
Can I suggest you register for an ORCID today? It will help you get credit for your work and move us towards more open science.
CC-BY
Article by Quentin Groom (NBGB)





 pro-iBiosphere wiki platform
pro-iBiosphere wiki platform
 RSS news
RSS news






 GBIO Info Sheet
GBIO Info Sheet


