22.04.2014
Hacking OCR for pro-iBiosphere
* by David P. Shorthouse, Rod Page, Kevin Richards, Marko Tähtinen
Taking his own lead from a pitch he delivered to an audience of receptive biodiversity informaticians at the outset of the March 17-21, 2014 pro-iBiosphere hackathon, Rod Page (University of Glasgow) fashioned an engaging interface to edit the OCR text from scanned pages in the Biodiversity Heritage Library (BHL). He wooed David P. Shorthouse (Canadensys), Kevin Richards (ex Landcare Research New Zealand) and Marko Tähtinen (University of Eastern Finland, BioVeL) away from eight other competing task groups, each of which issued products in a remarkably short amount of time.
The purpose of the pro-iBiosphere hackathon was to "enrich structured biodiversity input data with semantic links to related resources and concepts". The OCR task group led by Rod had a distinctly different starting point, one that is no less important to the semantic linking of biodiversity resources. The unstructured data in the BHL is arguably the richest source of freely accessible information for taxonomists and biodiversity enthusiasts that can be mined into structured data. However, the quality of its OCR output suffers from variable typefaces, layouts, page contrasts and page bleeding, artifacts and other issues that occasionally bewilder its OCR engine. As a result, data mining and indexing routines that lift scientific names, place names, and other entities in support of semantic linking are not always successful. The browsing interface in the BHL could be made more engaging if visitors had an opportunity to rapidly correct the OCR text while viewing the original scanned image, thus enriching search and discovery for future visitors. Indeed, BHL and its partners were recently awarded a "Digging Into Data Challenge" grant (see http://blog.biodiversitylibrary.org/2014/03/first-meeting-of-mining-biodiversity.html), part of which will employ automated text-cleaning methodologies developed by its Canadian collaborators. An OCR editor might complement their funded work. Likewise, the Finnish National Library has developed its own OCR editor interface (see http://blogs.helsinki.fi/fennougrica/2014/02/21/ocr-text-editor/). Unlike the Finnish editor that uses ALTO XML as its source documents, the OCR editing interface developed during this hackathon uses BHL’s DjVu XML documents as its source, rendered as HTML5.
The OCR Task Group had one aim: provide a simple interface for interactive editing of text, as well as tools to make inferences from the edits. After four solid days of hacking, the team completed this aim and integrated value-added features to engage users and to boost developer confidence in reuse of the code. The underlying document store is the cloud-based CouchDB (on Cloudant) and the team is confident that the proof-of-concept can be made to scale. The capabilities of the software are:
-
An in-place panel shows the exact line in the original scanned image while the user edits a single line of OCR text at a time (Figure 1)
-
Global Names scientific name-finding is integrated in real-time when a user completes a line edit, giving feedback if a scientific name is newly recognized (Figure 2)
-
Authentication uses the facile https://oauth.io/ such that all edits are tied to users’ OAuth2-provider accounts (eg Google, Twitter, GitHub)
-
Frequencies of common edits are summarized in real-time and other words that may benefit from similar edits are highlighted for users
-
Batch processes collapse all user edits and text files are recreated for possible re-introduction into data mining routines
-
Unit and integration tests are included
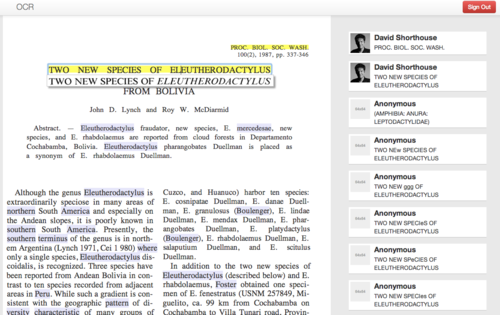
Figure 1. The OCR Editing interface rendered as HTML5, illustrating the original line of text as a clipped image under the line being edited, a scrolling tally of user edits, lines that have been previously edited (yellow highlight) and words that share strings of characters that match previous edits elsewhere on the page (mauve highlight).
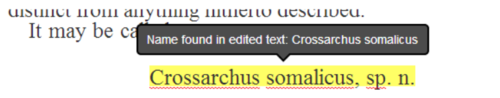
Figure 2. Tooltip showing a scientific name newly recognized by the Global Names Recognition and Discovery service when a user completes an edit.
A proof-of-concept can be examined at http://bionames.org/~rpage/ocr-correction/ and the MIT-licensed code can be obtained from https://github.com/rdmpage/ocr-correction. The team will follow-up with the BHL to share what was accomplished and to discuss how this could be integrated in their web-based interface.
The team spent the last day of the hackathon investigating the production of DjVu XML files from scanned specimen labels. Although investigations are still underway, this particular outcome would be an excellent enhancement to the workflow at the Université de Montréal Biodiversity Centre (David P. Shorthouse) and useful for other members of the Canadensys network. The OCR editing interface may also be useful for the multi-national Notes From Nature crowd-sourcing initiative, http://www.notesfromnature.org/ as well as other national, regional, and local specimen label digitizing efforts throughout the world.
Rutger Vos and Soraya Sierra (Naturalis, co-organizers) received abundant praise by all participants at the completion of the hackathon, and rightly so. The hackathon was exceptionally well organized, developer team sizes were perfect for each of the nine task groups, each participant’s work ethic was remarkable, facilities were well provisioned, and nibbles and luncheons were delectable. We look forward to the reactions of pro-iBiosphere members at the final event in Meise, Brussels.
Contact:
David P. Shorthouse
Université de Montréal Biodiversity Centre / Canadensys, Montréal, QC CANADA
Email: [email protected]





 pro-iBiosphere wiki platform
pro-iBiosphere wiki platform
 RSS news
RSS news
 Print this article
Print this article
